Autophagy
Autophagy is a cellular self-digestion process for the purpose of providing nutrients to allow for cell survival during stress conditions. Autophagy can be selective (mitophagy) or non-selective (nutrient poor). Autophagy plays an important role in maintaining the cell homeostasis by removing non-essential cell components. The role of autophagy is generally understood to be securing nutrient sources through autodigestion in order to survive starvation conditions. However, in recent years, it has been found to work along with the proteasome system in the metabolism of cell components even in normal environments. Compared to proteasomes, which selectively degrade ubiquitinated proteins as their targets, in autophagy the space within the cell is entirely digested; therefore it is called a bulk degradation system. Recent researches and advances have shown an association of mammalian autophagy with diseases such as neurodegenerative disease, infection disease, cardiac disease as well as cancers.
Development of autophagy research
Autophagy was first discovered in yeast as a form of cellular self-digestion for the purpose of providing nutrients to the cell in order to survive starvation. Research in autophagy has been expanding from its initial discovery in yeast, as a form of cellular self digestion, to having implications in major drug discovery pipelines. Recent researches and advances have shown an association of mammalian autophagy with diseases such as neurodegenerative disease, infection disease, cardiac disease as well as cancers.
What is Autophagy?
Autophagy is generally considered as a process to supply nutrients by self-digestion for cells to survive starvation. However, autophagy, along with the proteasome system, is also involved in the turnover of cellular components under normal conditions.
While proteasomes target and selectively degrade ubiquitinated proteins, autophagy degrades all the contents engulfed by autophagosomes, and, therefore, is called “the bulk degradation system.” In addition, selective autophagy pathways target cellular organelles, such as mitochondria and peroxisomes. These degradation mechanisms are respectively known as “mitophagy” and “pexophagy.” Various other autophagic mechanisms are also under investigation.
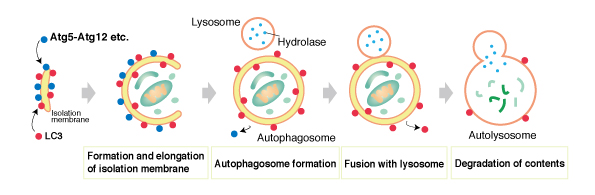
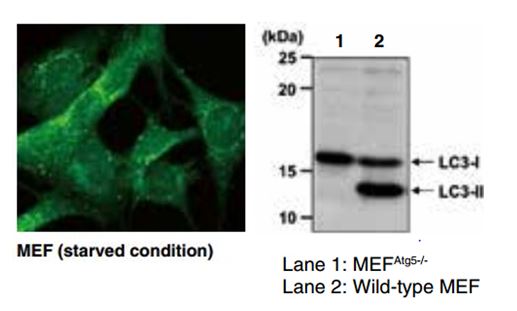
When a cell is placed under starvation conditions, a flat vesicle called an isolate membrane appears in the cytoplasm (1). Subsequently, the membrane extends while taking in the cytoplasm (2), its edges fusing to compose an autophagosome (AP) (3). Mitochondria and other large organelles are also contained within the AP. When the AP fuses with a lysosome (4), its contents are degraded (5). The amino acids gained through autodigestion are reused as nutrient sources. It is not known at this time how the isolate membrane appears or how its components are obtained.
Although in the limelight in recent years, autophagy was first observed by electron microscopy over 40 years ago. Nevertheless, functional studies of autophagy did not progress rapidly because factors involved in the process remained unknown for a long period of time.
Dr. Yoshinori Ohsumi (currently of the Tokyo Institute of Technology) and his colleagues at the National Institute for Basic Biology isolated yeast strains that were unable to degrades the contents of autophagosomes, and successfully cloned the autophagy-related (APG/ATG) genes (Tsukada and Ohsumi, 1993). As of 2016, the number of ATG genes in budding yeast stands at 41. Many of these genes are conserved in mammals and plants (the amino acid sequence homology among species is limited, but the 3D structures are similar).
With the discovery of APG/ATG genes, functions of the gene products have been extensively studied, and details of the mechanism and physiological role of autophagy are being elucidated one after another.
What is Selective autophagy?
Selective autophagy
Autophagy was initially thought to be a non-selective degradation mechanism, because the entire vesicle contents were digested. However, recent findings have revealed the selective degradation of mitochondria and other specific organelles, bacteria, and aggregates of proteins with attached ubiquitin chains (polyubiquitinated proteins). This mechanism is called “selective autophagy.”
Selective autophagy and p62
Because autophagosomes do not exhibit selectivity, “adaptor proteins” are necessary to link autophagosomes to proteins destined for selective degradation. One of these adaptor proteins is p62.
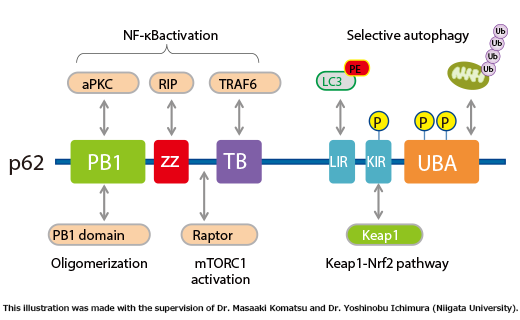
62/SQSTM1 is a scaffolding protein that interacts with various signaling molecules such as TRAF6, RIP, and aPKC (Fig. 1).
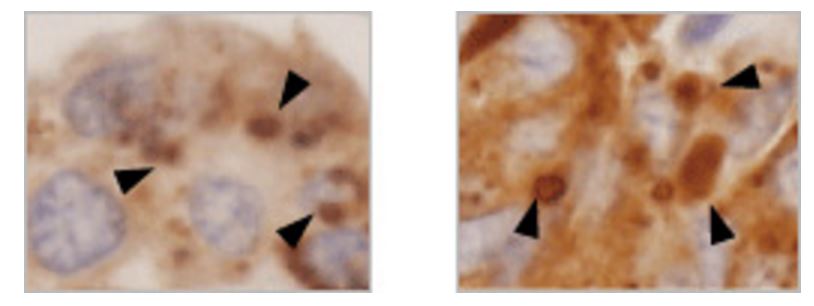
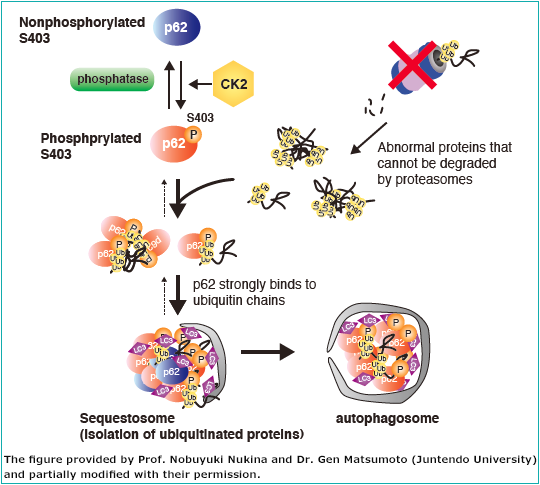
p62 contains an LC3-interacting region and is believed to be a substrate for selective autophagy. In addition, p62 contains a domain that binds ubiquitin chains, and mediates the recruitment of poly ubiquitinated protein aggregates and depolarized mitochondria to the autophagic machinery (see page 11 for the details of selective autophagy). In fact, in liver- and brain-specific autophagy-deficient mice, overaccumulation of p62 occurs, and ubiquitin- and p62-positive inclusion bodies are observed (Fig. 2). Importantly, ubiquitin- and p62-positive inclusion bodies are also observed in tissues of patients with neurodegenerative diseases (such as Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis), alcoholic hepatitis, hepatic steatosis, and liver cancer. There is increasing interest in the involvement of impaired autophagic degradation of p62 in these diseases.
p62/SQSTM1 domain structure (Fig. 1)
Selective autophagy of poly ubiquitinated protein by p62 (Fig. 2)
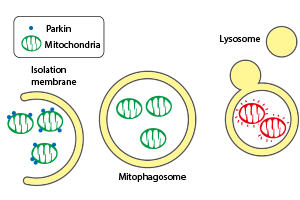
Mitophagy is a type of autophagy that selectively degrades mitochondria, and is involved in the turnover of damaged mitochondria. This process is thought to defend the body from diseases resulting from mitochondrial dysfunction. The Parkinson’s disease gene product, Parkin (ubiquitin ligase), plays a critical role in the induction of mitophagy. Parkin is recruited to the outer membrane of damaged and depolarized mitochondria. Ubiquitin is subsequently added to the outer membrane of damaged mitochondria by the ubiquitin ligase activity of Parkin. Mitophagy is induced through the recognition of the ubiquitin modification.
Phospho-p62: Hot topic in research on neurodegenerative disease and cancer
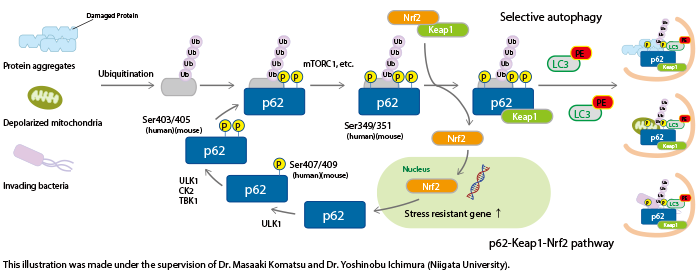
p62 contains multiple phosphorylation sites. Sequential phosphorylation of these sites regulates biological defense mechanisms such as selective autophagy.
The phosphorylation of Ser407 (human)/Ser409 (mouse) precedes the phosphorylation of Ser403 (human)/Ser405 (mouse) in p62, which increases its affinity for poly ubiquitin chains. Consequently, ubiquitinated abnormal protein aggregates, depolarized mitochondria, and invading intracellular bacteria are sequestered by phospho-p62. Further phosphorylation of Ser349 (human)/Ser351 (mouse) by mTORC1 increases the affinity of p62 for Keap1, inducing dissociation of Nrf2 from Keap1 and nuclear translocation of Nrf2 (the p62-Keap1-Nrf2 pathway). Nrf2 is a stress-response transcription factor and activates the transcription of various stress resistance genes. Nrf2 also induces p62 gene expression, forming a positive feedback loop. Phospho-p62 with bound Keap1 interacts with LC3 through the LIR (LC3-interacting region) and is degraded by the autophagy pathway. Thus, the cells under stress conditions effectively overcome their negative environment by activating two biological defense mechanisms through the phosphorylation of p62.
Impaired selective autophagy is implicated in various diseases. For example, neurons in familial parkinsonism fail to clear protein aggregates and depolarized mitochondria, resulting in neuronal damage and compromised brain function. In hepatocarcinoma cells, p62 is constitutively phosphorylated at Ser349, causing continuous activation of Nrf2. Hence, inhibitors of p62 phosphorylation and inhibitors of the interaction between phospho-p62 and Keap1 have the potential to be novel cancer therapeutics.
(Reference: Saito, T., et al., Nat. Commun. 7, 12030 (2016) PMID: 27345495).
The p62-Keap1-Nrf2 pathway
Atg family proteins
The discovery of APG/ATG genes
Although in the limelight in recent years, autophagy was first observed by electron microscopy over 40 years ago. Nevertheless, functional studies of autophagy did not progress rapidly because factors involved in the process remained unknown for a long period of time.
Dr. Yoshinori Ohsumi (currently of the Tokyo Institute of Technology) and his colleagues at the National Institute for Basic Biology isolated yeast strains that were unable to degrades the contents of autophagosomes, and successfully cloned the autophagy-related (APG/ATG) genes (Tsukada and Ohsumi, 1993). As of 2016, the number of ATG genes in budding yeast stands at 41. Many of these genes are conserved in mammals and plants (the amino acid sequence homology among species is limited, but the 3D structures are similar).
With the discovery of APG/ATG genes, functions of the gene products have been extensively studied, and details of the mechanism and physiological role of autophagy are being elucidated one after another.
5 complexes which are involved in autophagosome formation
The 17 factors necessary for autophagosome formation which are marked with red circles above can be classified to 5 functional groups below;
- Atg8-conjugation system: Atg8, Atg4, Atg7, Atg3
- Atg12-conjugation system: Atg12, Atg7, Atg10, Atg5, Atg16
- Atg1 protein kinase complex: Atg1, Atg13, Atg17, Atg29
- The Vps34 PI3 kinase complex: Vps34, Vps15, Atg14, Atg6
- Atg9 and Atg2-Atg18 complex : Atg2, Atg18, Atg9
Atg proteins and their functions
| Yeast | Mammalian | Functions |
|---|---|---|
| Atg1 | ULK1, 2 | Atg1/Atg13/Atg17/Atg29 kinase complex |
| Atg2 | ATG2A, ATG2B | Atg2/Atg18/Atg9 complex (autophagosome formation) |
| Atg3 | Atg3 | E2-like enzyme specific for Atg8 (autophagy induction) |
| Atg4 | Atg4A, 4B, 4C, 4D | Cysteine protease: Cleavage of C-terminal of Atg8 and de-PE |
| Atg5 | Atg5 | Atg12/Atg5/Atg16 complex (autophagosome formation) |
| Atg6/Vps30 | Beclin-1 | A subunit of Vps34 PI3K complex (autophagosome formation) |
| Atg7 | Atg7 | E1-like enzyme: Both Atg8 and Atg12 are Atg7 substrates |
| Atg8 | LC3, GABARAP, GATE-16 | Ubiquitin-like protein: Atg8-PE formation (phosphatidylethanolamine) |
| Atg9 | ATG9A(APG9L1) TTG9B(APG9L2) |
Atg2/Atg18/Atg9 complex (autophagosome formation) |
| Atg10 | Atg10 | E2-like enzyme specific for Atg12 (autophagy induction) |
| Atg11 | Adaptor molecule: Incorporation of API into Cvt vesicle in yeast | |
| Atg12 | Atg12 | Ubiquitin-like protein: Atg12/Atg5/Atg16 complex |
| Atg13 | Atg13 | A subunit of Atg1 complex (autophagy induction) |
| Atg14 | ATG14, ATG14L, BARKOR | A subunit of Vps34 PI3K complex (autophagy induction?) |
| Atg15 | Lipase-like protein: degradation of autophagic body in yeast | |
| Atg16 | ATG16L1, ATG16L2 | Atg12/Atg5/Atg16 complex (autophagosome formation) |
| Atg17 | A subunit of Atg1 kinase complex (autophagosome formation) | |
| Atg18 | WIPI-1, 2 | PIP3 binding protein: Atg9/Atg2/Atg18 complex |
| Atg19 | The receptor of API in Cvt pathway (yeast) | |
| Atg20 | PI3P binding protein in Cvt pathway (yeast) | |
| Atg21 | WIPI-1, 2 | PI3P binding protein in Cvt pathway (yeast) |
| Atg22 | A membrane protein of vacuoles in yeast | |
| Atg23 | Cvt vesicle formation in yeast | |
| Atg24 | Cvt pathway and degradation of peroxisome (yeast) | |
| Atg25 | Degradation of peroxisome in yeast | |
| Atg26 | Degradation of peroxisome in yeast | |
| Atg27 | PI3P binding protein in Cvt pathway (yeast) | |
| Atg28 | Degradation of peroxisome in yeast | |
| Atg29 | A subunit of Atg1 kinase complex (autophagy induction?) | |
| Atg30 | Peroxisome degradation (Pexophagy) | |
| Atg31 | Atg17-Atg29-Atg31 complex in starvation induced autophagy | |
| Atg32 | BCL2L13 | Mitochondria degradation (Mitophagy) |
| Atg33 | Mitochondria degradation (Mitophagy) | |
| Atg34 | Alpha-mannosidase transport | |
| Atg35 | Peroxisome degradation (Pexophagy) | |
| Atg36 | Peroxisome degradation (Pexophagy) | |
| Atg37 | ACBD5 | Acyl-CoA binding protein for forming isolation membrane. |
| Atg38 | Interacts with Atg14 and Vsp34. Plays a role as a linker between Vps15-Vsp14 and Vsp30/Atg6-Atg14. | |
| Atg39 | Selective autophagy of the nuclear membrane. | |
| Atg40 | Selective autophagy of the endoplasmic reticulum. | |
| Atg41 | Interacts with Atg9, and is involved in PAS formation. |
Reference: Klionsky DJ et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 12, 1-222, 2016 (PMID: 26799652)
Atg8-conjugation system
After the ubiquitin-like protein Atg8 has had its C-terminal arginine residue cleaved by the cysteine protease Atg4, it is passed on to E1 (Atg7) and E2 (Atg3) and transferred into the head group of its substrate phosphatidylethanolamine (PE). This Atg8-PE conjugate functions as part of the membrane component of the autophagosome. When Atg8-PE is once again deconjugated PE by Atg4, the Atg8 is recycled. Incidentally, an E3-like protein in autophagy has not yet been found.
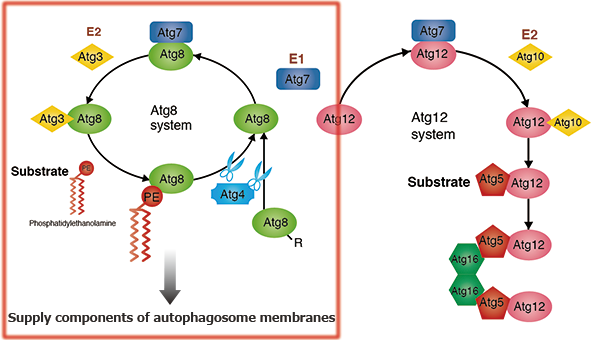
Atg12-conjugation system
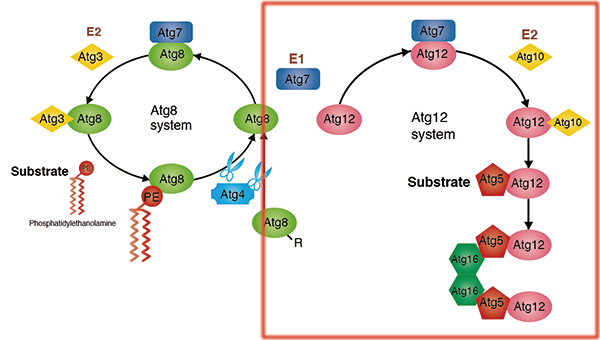
Unlike other ubiquitin-like proteins, the ubiquitin-like protein Atg12 has a C-terminal glycine, which protects it from processing. Atg12 is conjugated to the substrate Atg5 by Atg7 (an E1-like protein, and as seen in the conjugation of Atg8 to PE) and Atg10 (an E2-like protein). The Atg12-Atg5 conjugate forms a complex with Atg16. Self-oligomerization of Atg16 results in a multimer of the Atg12-Atg5-Atg16 complex. Although its functions remain unknown, this complex is shown to accumulate on the pre-autophagosomal structure (PAS) in yeast and to play an essential role in autophagosome formation.
Atg1 protein kinase complex
Induction of autophagy is mediated by inactivation of the serine/threonine protein kinase known as target of rapamycin (TOR). Among other molecules, Atg13 is highly phosphorylated by the TOR under nutrient-rich conditions, being unable to bind to Atg1. In such conditions, the kinase activity of Atg1 is kept low as well.
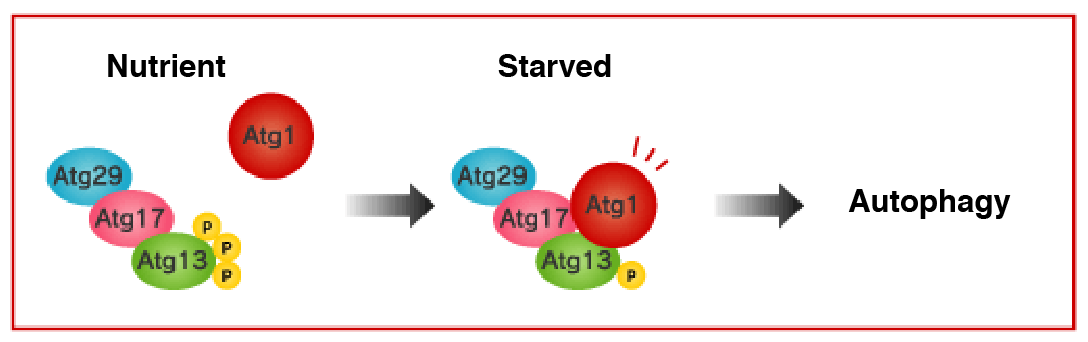
In contrast, under starvation conditions, TOR is inactivated and the phosphorylation levels of Atg13 decrease. Then, Atg13 and Atg17 bind to Atg1, and kinase activity of Atg1 also increases. Induction of autophagy is known to be dependent on this kinase activity of Atg1, but further researches are necessary to fully understand its detail. Rapamycin, a TOR inhibitor, also induces the same phenomenon. It has been shown that Atg29 binds to Atg17 in the budding yeast.
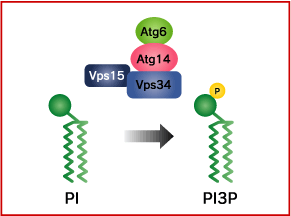
The Vps34 PI3 kinase complex
Phosphatidylinositol 3-kinase (PI3K) phosphorylates the phosphatidylinositol (PI) in the membrane lipid to create phosphatidylinositol 3-phosphate (PI3P). Class III PI3K activity, called Vps34, is necessary for the progress of autophagy.
Vps34 forms a complex with Vps15 (protein kinase), Vps30/Atg6 (Beclin-1 in mammals), and Atg14. If even one of these subunits is rendered defective, autophagy will not be induced. As well, because lipid kinase activity in Vps34 is necessary for autophagy to take place, it is thought that the PI3P produced thereby plays some role in autophagy. Beclin 1/hVps34/p150/Rubicon/UVRAG complex involves in autophagosome maturation. On the other hand, Beclin 1 has been reported to be defective in various forms of cancer (breast, ovarian, prostate), and it is known that cancer is frequent in Beclin 1 hetero-defective mice. As well, DNA defects related to cancer have been found in the Beclin 1 binding protein UVRAG.
Atg9 and Atg2-Atg18 complex
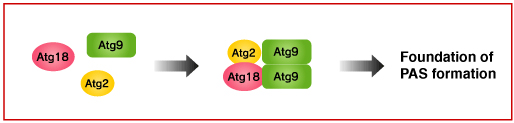
It has not yet known where the initiation of autophagy takes place. In the yeast, however, autophagosome formation is suggested to start at the pre-autophagosomal structure (PAS). Atg9 is thought to be involved in the early phase of PAS formation, since many Atg proteins are not localized in the PAS in the budding yeast lacking ATG9.
Atg2-Atg18 complex binds to Atg9 on the surface of the PAS and Atg18 is known to interact with PI3P. But, the function of Atg2-Atg18 complex is still not known. In the yeast with mutated ATG18 and ATG2, Atg9 is highly accumulated at the PAS. Meanwhile, Atg9 is not localized at the surface of autophagosome. These findings suggest there must be mechanisms for Atg9 to detach from the PAS surface. Atg2-Atg18 complex may be involved in this process.
Factors related to autophagy continue to increase
As we have seen so far, it is now understood that the seventeen varieties of Atg proteins necessary for the formation of an autophagosome act as at least five functional aggregates. There are many issues yet to be understood regarding the role of each complex and the interactions among complexes, but in recent years more and more factors related to autophagy, beginning with Atg conjugated proteins, continue to be identified.
UVRAG, related to UV resistivity, is receiving renewed attention as a Beclin 1 conjugated protein, and it is now clear that it is involved in the formation of the autophagosome through the activation of PI3K. p62/a170/SQSTM1 (p62), which interacts with Atg8 (LC3), is a scaffolding protein for signal transmission through ERK or PKC; because it has a ubiquitin binding domain at its C-terminal, it suggests crosstalk between ubiquitin proteasome degradation through p62 and autophagy. As well, regarding pathogen elimination through autophagy, it is known that LRGM/LRG-47, a variety of GTP conjugated protein with the toll-like receptor and IFN-γ inductor important for pathogen recognition, is used in innate immunity.
In this way, spurred by research into autophagy, new and diverse interactions among biological phenomena have become visible.
In the future, expectations will rest on approaches using Atg-factor defective yeast strains and Tg/KO mice, as well as development of factor monitoring tools such as antibodies to Atg proteins and conjugate factors, and fluorescent protein fusion proteins.
Autophagy Products Highlights
Antibodies against p62/SQSTM1
Detection of p62-positive inclusion bodies in the human hepatocellular carcinoma using PM045