The Principle and Method of Immunoprecipitation (IP)
Contact Us for help in setting up your experiments
The principle
Immunoprecipitation (IP) is a method to isolate a specific antigen from a mixture, using the antigen-antibody interaction. Antigens isolated by IP are analyzed by SDS-PAGE or Western blotting.
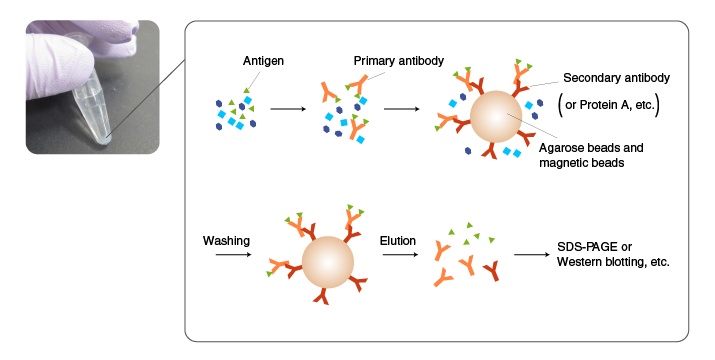
In IP, an antibody is added first to a mixture containing an antigen, and incubated to allow antigen-antibody complexes to form. Subsequently, the antigen-antibody complexes are incubated with an immobilized antibody against the primary antibody (secondary antibody) or with protein A/G-coated beads to allow them to absorb the complexes. The beads are then thoroughly washed, and the antigen is eluted from the beads by an acidic solution or SDS.
If a suitable antibody is not available, the target molecule is fused to a His tag or other tags by recombinant DNA techniques, and immunoprecipitated using an antibody to the tag (pull-down assay).
The use of an antibody with high binding specificity and affinity for the antigen is critical for successful IP. Antibodies raised against synthetic peptides and recombinant proteins often work well in Western blotting but may not bind the antigens in their native conformation in solution. When using commercial antibodies, select the ones that are suitable for IP according to product information. Also recognize the properties of both the antibody and antigen in the literature and product information.
Tips for efficient IP
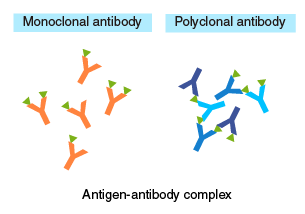
The diagram on the right illustrates the structures of antigen-antibody complexes formed with a monoclonal versus polyclonal antibody as the primary antibody. When using a monoclonal antibody as the primary antibody, adjust the concentration so that:
[Secondary antibody] > [Primary antibody] > [Antigen].
An excess of primary antibody, relative to the secondary antibody, may compete with antigen-antibody complexes for the secondary antibody, resulting in a lower yield of recovery. When using a polyclonal antibody, an excess of primary antibody, relative to the antigen, will prevent the formation of oligomeric complexes. Therefore, these concentrations should be optimized by testing different ratios.
Comparison of primary antibodies for IP
| Monoclonal Antibody | Polyclonal Antibody | |
|---|---|---|
| Background | Background is low with an appropriate antibody because the antibody recognizes a single antigen. | Background may be high if some of the antibody molecules have a low specificity and bind to proteins other than the target protein. |
| Binding affinity | High-affinity monoclonal antibody (dissociation constant Kd<10-8 M) should be used because low affinity antibody may not form an antigen-antibody complex in solution. | Even if the affinity of individual antibody molecules is low, oligomeric antigen-antibody complexes are formed easily due to the multivalent binding. |
| Stability of antigen-antibody complexes | If the binding affinity of an antibody is low, simultaneously using several high-specificity monoclonal antibodies will allow multivalent binding, resulting in stable antigen-antibody complexes | Stable oligomeric complexes are formed because reaction between a polyclonal antibody and an antigen is multivalent. |
Beads for immobilization
Agarose beads and magnetic beads are commonly used. Agarose beads have a porous, mesh-like structure, and antibodies can diffuse and bind to the internal matrix of the beads, which provides high binding capacity. Magnetic beads are simple spheres, providing ease of handling and short processing time. With appropriate coating, background can be reduced. However, binding capacity may not be high enough for some applications, and the cost is higher than the alternative using agarose beads.
Magnetic agarose beads provide ease of handling (when used with a magnet) and a high binding capacity.
Description of Each Conjugate
Detergents
Appropriate concentrations of salt and non-ionic detergent are commonly used in IP to reduce non-specific binding (protein-protein and protein-bead interactions). Pilot experiments should be performed with detergents, which may reduce the affinity of the antibody, especially when using a monoclonal antibody.
Protease inhibitors
The target protein and antibody are subject to degradation by protein-degrading enzymes (proteases) in samples such as cell lysates and tissue extracts. To prevent proteolytic degradation, protease inhibitors are included. When the type of proteases in samples are known or predicted, specific inhibitors are used. When proteases are unknown, a combination of multiple small molecule inhibitors, such as PMSF and EDTA, are used.
Elution condition
In most applications, proteins are eluted in SDS sample buffer containing a reducing agent, such as 2-mercaptoethanol (2-ME). In addition to the target protein, the antibodies used for IP are co-eluted, which should be considered when performing Western blotting or mass analysis.
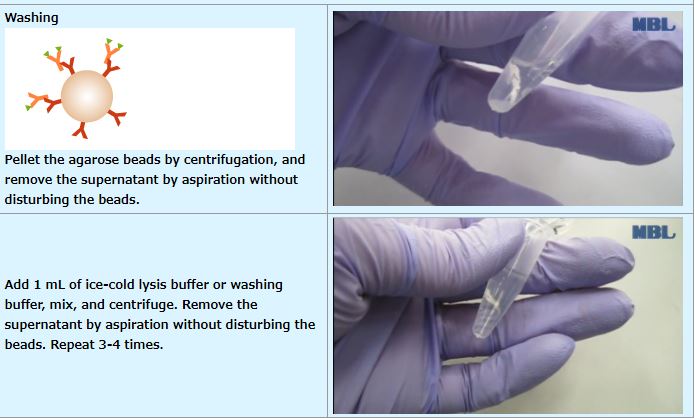
Procedure
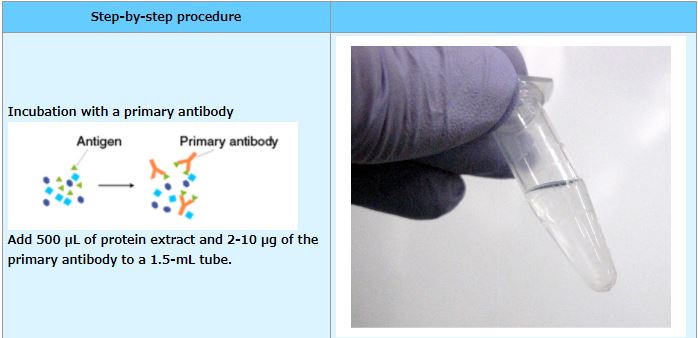
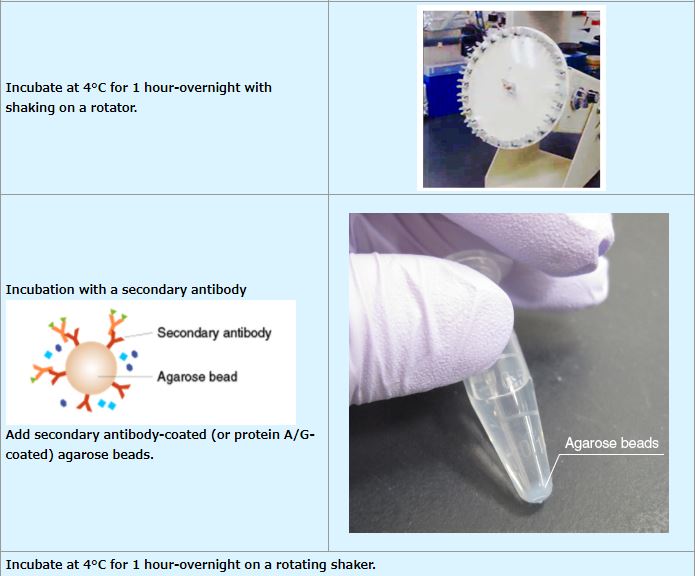
IP (with agarose beads)
An example performed at MBL

Previous page: Enzyme-linked Immunosorbent assay (ELISA) | Next page: Co-Immunoprecipitation (Co-IP)
Related Links
Antibody
Antibody basics
Antibodies as a research tool
- How to generate antibodies
- How to select antibodies
- Labeled antibodies
- How to label antibodies
- Main causes of non-specific reactions
- How to reduce non-specific reactions
- Tags and Tag antibodies
Qualitative and quantitative measurements of proteins using antibodies
- Western blotting (WB)
- Enzyme-linked Immunosorbent assay (ELISA)
- Immunoprecipitation (IP)
- Co-immunoprecipitation (Co-IP)